The Nobel Prize-winning discovery and invention of the polymerase chain reaction (PCR) in the mid-1980s has allowed biologists to design very specific DNA markers to assess the presence or absence of certain genes.
Generating marker information from a number of individuals, using markers on all chromosomes, allows the development of a DNA fingerprint unique to each individual (be they human, animal, plant or microbe). The most complete DNA fingerprint of any individual is the full genome sequence – every single nucleotide base (A, C, G and T) on all the chromosomes. For tomatoes, this works out to 950-million bases! This level of detail is not always required for breeding, and more specific DNA markers that target genes of interest are used for day-to-day research.
Plants are attacked by a whole suite of plant pathogens: viruses, bacteria, fungi, nematodes, etc. In some individuals, resistance genes stop the pathogen from infecting, or limit its spread and the damage that it causes. There are numerous types and classes of disease resistance genes, which differ in their functioning mechanism. Chemical spraying can limit crop losses when pathogens infect, but disease resistance presents a simpler, cheaper and often more robust way of protecting a crop. In many instances, chemical control is very limited (e.g. bacterial infections) or not possible (e.g. against most viruses), so varieties with resistant genes are a farmer’s best hope.
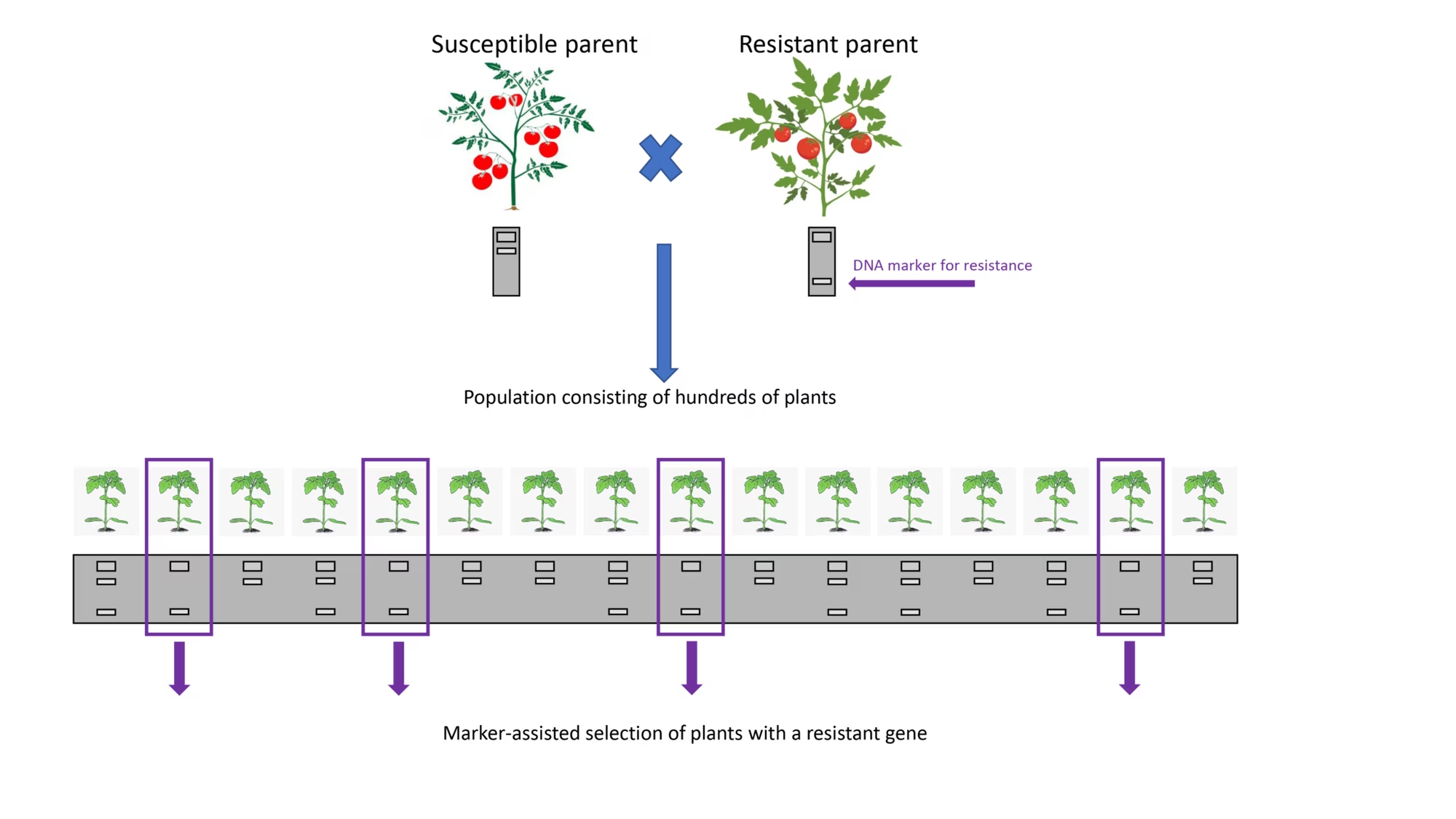
When an individual plant or wild crop relative has a resistance gene, it is transferred through cross-pollination into domesticated varieties to be used in a breeding programme. But unless the pathogen is present and conditions favourable in the field during that season, it’s possible that infection will not occur, and a breeder will not be able to select resistant plants because they’ll all look resistant.
Scientists have found a way round this uncertainty by mapping the exact chromosomal location of resistance genes, and then using a DNA marker to test if a plant contains or lacks that specific gene (resistant or susceptible, respectively). Being able to evaluate the genetic combination of plants in this way is known as marker-assisted selection (MAS), and has a number of benefits.
First, the process of genotyping is rapid (as short as a few days in the lab), compared to a long period of disease development required for some pathogens. Second, environmental conditions that lead to uneven infection in the field don’t affect the ability of a DNA marker to distinguish resistant from susceptible individuals, as the DNA remains stable. Third, leaf samples from a breeding population can be taken at a seedling stage, and specific seedlings containing the resistance gene only can be selected for transplanting, saving time, space and money. Fourth, plants can be tested for multiple traits or resistance genes at one time compared to artificial inoculation, where only a single pathogen can be used to test lines.
MAS has become such a vital part of the breeding process that now, many external service laboratories offer the option of running DNA markers for disease resistance genes on a set of leaf or seed samples. Because many diverse breeding lines can be combined together in a population, dozens of different DNA markers can be used to find that rare plant in which a number of important genes are combined. These still need to be transplanted and grown to maturity to ensure the yield, fruit quality and size remain acceptable.
In some instances, incorporating disease genes from wild species brings with them some unwanted baggage – flanking DNA of the wild species that has a negative impact, such as smaller or softer fruit, lower yield or reduced plant vigour. This trade-off occurs often, and in many cases older varieties without disease resistance have higher yields and better fruit quality than newer varieties that are resistant to many pathogens. Sometimes, this “linkage drag” can be eliminated by repeated crossing to domesticated varieties combined with MAS, but in some cases it’s just unavoidable.
As COVID-19 has taught us, pathogens can mutate and change. When this happens in plant pathogens, they may become more aggressive and overcome the resistance genes that have been deployed. This starts a whole new cycle of evaluating wild relatives for new disease genes that are effective against the new strain of the pathogen, incorporating them into domesticated varieties, mapping the location of the new gene, and then using MAS to track the gene in a breeding population.
In Starke Ayres’ research, our team of breeders and biotechnologists actively look for new disease genes, map them to specific chromosomes and then pass this info on to our breeders. Combined across our crops, our breeding teams track over 50 different genes, and in 2021 we analysed over 60,000 leaf samples and generated 2,850,000 data points in our Biotechnology lab.
Our breeders use this data on a daily basis to make selections of superior plants that will go on to produce the next generation of resistant varieties for our customers.